The roles of alternate transcripts and uORFs in regulating protein synthesis in meiosis and beyond
(including Ünal lab collaborations)
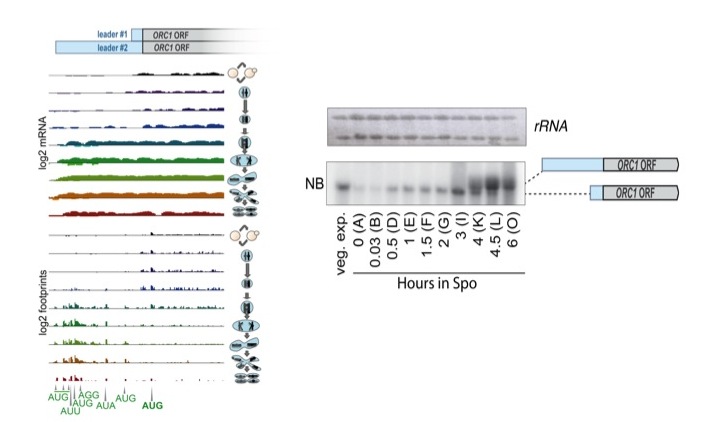
The layered setup of our initial survey of meiotic gene expression allowed us to observe the interplay between transcriptional and translational control, providing a rich view of the integration of gene regulation that underlies the complex cellular remodeling and chromosome segregation associated with meiosis. We identified an interesting example of such regulation for the conserved DNA replication factor, Orc1. Orc1, a member of the six factor Origin Recognition Complex, is required for both mitotic and meiotic DNA replication. In meiosis, a longer transcript of ORC1 is seen in the cell after meiotic DNA replication is completed. This longer transcript allows only poor translation of Orc1 protein while subject to efficient translation of several uORFs in the leader. These uORFs appear to compete with the ORC1 ORF for translation capacity, and reduce the amount of this essential factor at a time that meiotic cells no longer require it.
Orc1 protein synthesis is controlled by uORF-mediated translation competition on a regulated long meiotic transcript isoform. mRNA and ribosome footprint sequencing are shown for ORC1 , with mitotic cells in the top (black) track and meiotic progression proceeding from top to bottom below that. In mitotic and early meiotic cells, a short transcript is present that allows Orc1 translation and doesn't contain uORFs. Following meiotic translation, a longer transcript is observed, with several AUG uORFs translated that appear to prevent ribosomes from translating the ORC1 ORF.
We originally identified ~100 other examples of such regulation, often of important genes in driving the meiotic program. One particularly interesting example is the conserved kinetochore component, Ndc80, which is strongly translationally regulated in meiosis, though with very different kinetics than is seen for Orc1. Ndc80 translation is kept low early in meiosis through the presence of a longer transcript with competitive uORFs prior to the first round of meiotic chromosome segregation. When this regulation is disrupted, the meiotic pattern of chromosome segregation does not occur properly (Miller and Ünal et al., eLife, 2012). Elegant work by the Ünal and Van Werven labs showed that the mechanism of regulation responsible for setting Ndc80 protein levels involves regulated switching between two transcript isoforms with different transcription start sites (TSSes). Transcription from the upstream TSS produces a 5’ extended transcript isoform that is well translated for uORFs but not for the NDC80 ORF and whose production silences the downstream TSS in cis, through chromatin modifications (Chen and Tresenrider et al., eLife, 2017; Chia et al., eLife, 2017). This extended mRNA was named “LUTI” for long undecoded transcript isoform.
LUTI-based regulation commonly drives meiotic protein levels
Our lab’s matched measurements of mRNA, translation, and protein during meiosis (in collaboration with the Jovanovic lab) revealed that a large proportion of genes expressed during meiosis showed a peculiar anti-correlation between mRNA and protein levels over time (Cheng and Otto et. al., Cell, 2018). This type of relationship cannot be explained by traditional gene regulatory mechanisms, but we noted that both ORC1 and NDC80 fell into this category. We queried our dataset for genes with features common to the LUTI-based regulation determined for NDC80 and found 380 genes to be regulated by this mechanism, representing nearly 8% of all yeast genes for which we collected measurements.
LUTI-based regulation. Top) Anti-correlated mRNA and protein levels during meiosis (represented by cartoon) are explained by LUTI regulation. Here, TF1 induces a LUTI transcript early in meiosis, which results in high overall mRNA levels for this gene, but poor protein production due to translation of uORFs that repress ORF translation. In mid-meiosis, TF2 induces low levels of a canonical mRNA, which produces protein. Below) LUTI vs. and canonical mRNA production is determined by the TF abundances and the positions of their binding sites relative to ORF and uORF sequences. Not shown: transcription from TSS1 represses TSS2 transcription in cis, causing toggling from canonical to LUTI mRNA isoform. This mechanism regulates over 380 genes during meiosis in yeast.
LUTI-based regulation can result in an inverse correlation between mRNA and protein levels over time because 1) The LUTI transcript is often more abundant than the canonical transcript but its ORF is not efficiently translated, and 2) Toggling occurs between the two mRNA isoforms, such that an increase in the abundant LUTI results in a decrease in the translatable mRNA. By this regulation, traditional rules of gene regulation must be reconsidered. Although they contain ORF sequences, LUTI mRNAs do not seem to be intermediates between DNA and protein, and appear exist to repress production of the translatable isoform. Both transcriptional repression (cis-interference of the canonical TSS by transcription from the LUTI-producing TSS) and translational interference (repression of translation of the ORF by uORFs on the LUTI) are needed for this regulation to repress protein production from this locus. Either alone is not repressive for protein production.
As a result of this integrated regulation, a transcription factor (TF) that drives production of a LUTI mRNA is actually repressing protein production for the encoded ORF. The idea that driving more mRNA results in less protein is counterintuitive, but we proved this causality for meiotic genes regulated by the LUTI mechanism by forcing expression of a key meiotic TF, Ndt80 (please note coincidental similarity of this name to Ndc80 above), in meiotic cells arrested in pachytene. LUTI targets of Ndt80 were accurately predicted based on presence of its binding site near their distal TSS, and showed an increase in overall mRNA abundance but a decrease in protein abundance that depended on Ndt80 induction. Importantly, this experiment revealed regulatory logic to this unconventional regulatory strategy- it enables coordinated up-regulation of protein production from one set of targets (canonical transcripts) and down-regulation from another (LUTIs).
With the recent ability to quantify gene expression globally at multiple levels, there has been intense interest in ascertaining the relative importance of different stages of gene regulation. Our study suggests that a focus on relative quantitative contributions may cause us to miss important qualitative changes. By the LUTI mechanism, timed changes in the transcript pool composition for a large set of genes, rather than their levels, are key in driving the changing composition of the proteome through cellular differentiation. A single transcription factor can activate protein expression or repress protein production, a distinction based not on whether an mRNA is induced, but on the position of the TSS relative to the ORF start codon and the resultant translatability of the specific isoform induced. This provides an elegant solution to the problem of precisely coordinating increases and decreases in protein expression during a developmental program. LUTI-based regulation precludes the need for a dedicated trans-factor for transcriptional repression and instead allows repurposing of existing transcription factors for a new function, dependent only on cis-sequence evolution.
LUTI-based regulation contributes to the canonical Unfolded Protein Response
Canonical UPR activation involves transcriptional up-regulation of both canonical and LUTI targets. Canonical targets are translated, resulting in protein accumulation and include classical UPR targets, like BiP (KAR2 in yeast). LUTI targets are bound by Hac1 at a distal TSS, upstream of uORF sequences and thus do not result in protein production. In fact, the production of these Hac1-dependent LUTIs causes shutdown of transcription from the proximal TSS and ultimately protein-level down-regulation of a group of genes enriched for roles in aerobic respiration.
A transcription-factor(TF)-driven cellular state change that is of interest to our lab (discussed below in ongoing projects for its relevance to meiosis) is the Unfolded Protein Response (UPR). In the central branch of the UPR, which is conserved from yeast to man, the TF Hac1 mediates a well-studied transcriptional response, involving up-regulation of genes including chaperones and those required for expansion of the endoplasmic reticulum (ER). We decided to test whether the LUTI-based transcript toggling mechanism that we had found to be common in meiosis1 was also a part of this pathway. In other words, we tested whether Hac1 induced any 5’ extended mRNA targets that resulted in protein down-regulation. After UPR induction by treatment with either DTT or tunicamycin, two classic exogenous activators of this pathway, we collected parallel measurements of mRNA, translation, and protein from matched extract of cells with or lacking Hac1. We found that, indeed, Hac1 has LUTI targets (Figure 2) and induction of these targets as part of the UPR drives down-regulation of respiratory chain proteins, mediating a shift away from aerobic respiration in UPR-activated cells that is similar to the Warburg effect, by which cancer cells favor glycolysis. Importantly, we showed that this shift is important for UPR function, as cells that were unable to respire were better at adapting to protein folding stress than wild-type cells.
This work was significant for two reasons. First, Hac1 is a well-studied TF and previous studies had annotated its transcriptional targets based on the assumption that mRNA increases cause protein increases. Some of its LUTI targets had been inadvertently mischaracterized as canonical translated targets based on this rational assumption, suggesting that other well-studied TFs might have LUTI targets that have been misidentified as positive targets. Second, by simply determining which genes were “negative” (LUTI) targets of Hac1, we identified a specific biologically coherent feature of this pathway that is important for its function. This study suggested that LUTI-based regulation might be used as a general strategy for coordination of up- and down- regulation during cellular transitions.
Features of LUTI-based regulation are seen for the human oncogene MDM2
The key features of LUTI-based regulation—alternate 5’ extended transcripts, transcriptional interference between distal and proximal TSSes, and competitive uORF translation—have all been seen to occur in mammalian cells, although they have not previously been studied as a set of three linked features in that context. We therefore investigated the possibility that LUTI-type of regulation might occur in mammals, or more precisely, that there might be mammalian genes for which these three features are all linked. In collaboration with the Ünal lab, as well as the Tjian and Darzacq labs, we led a study that focused on the oncogene MDM2, which had been shown in human cells to be encoded by two transcript isoforms, one of which is 5’ extended and contains uORFs that dampen translation of the ORF. Ina showed that CRISPRi-based inhibition of the distal TSS in human cell lines resulted in up-regulation of the proximal TSS in a dose-dependent manner and that this was associated with the local loss of H3K36 tri-methylation marks, which is required for the LUTI-based regulation defined in yeast. In other words, simply inhibiting the LUTI-like promoter caused up-regulation of the promoter for the well-translated transcript, suggesting that transcriptional interference occurs at this locus and that a function of the distally encoded transcript isoform might be to control regulation of the well-translated isoform. Further, we found regulated toggling between the two MDM2 isoforms during both neural and endoderm human embryonic stem cell differentiation programs, suggesting a natural developmental significance to this regulation.
Open questions
Although we have found this unconventional regulation involving regulated transcript toggling to be common and important during yeast meiosis and beyond, there are many outstanding mysteries. First, we don’t know how common this regulation is in non-yeast eukaryotes. Second, we don’t understand the rules for uORF-mediated repression on LUTI transcripts. Third, we don’t know the contribution of mRNA stability. All are areas of interest to our lab.